Provides a number of modifications to the plot that are necessary for relative abundance plots of a number of species. See scale_x_abundance, facet_grid, facet_grid, label_species, label_geochem, and rotated_facet_labels rotated_axis_labels for examples of how to customize the default behaviour.
facet_abundanceh(
taxon,
grouping = NULL,
rotate_facet_labels = 45,
labeller = label_species,
scales = "free_x",
space = "free_x",
dont_italicize = c("\\(.*?\\)", "spp?\\.", "-complex", "[Oo]ther"),
...
)
facet_abundance(
taxon,
grouping = NULL,
rotate_facet_labels = 0,
labeller = label_species,
scales = "free_y",
space = "free_y",
dont_italicize = c("\\(.*?\\)", "spp?\\.", "-complex", "[Oo]ther"),
...
)
facet_geochem_wraph(
param,
grouping = NULL,
rotate_axis_labels = 90,
scales = "free_x",
labeller = label_geochem,
renamers = c(`^d([0-9]+)([HCNOS])$` = "paste(delta ^ \\1, \\2)", `^210Pb$` =
"paste({}^210, Pb)", `^Pb210$` = "paste({}^210, Pb)"),
units = character(0),
default_units = NA_character_,
...
)
facet_geochem_wrap(
param,
grouping = NULL,
scales = "free_y",
labeller = label_geochem,
renamers = c(`^d([0-9]+)([HCNOS])$` = "paste(delta ^ \\1, \\2)", `^210Pb$` =
"paste({}^210, Pb)", `^Pb210$` = "paste({}^210, Pb)"),
units = character(0),
default_units = NA_character_,
...
)
facet_geochem_grid(
param,
grouping = NULL,
rotate_axis_labels = 0,
scales = "free_y",
space = "fixed",
labeller = label_geochem,
renamers = c(`^d([0-9]+)([HCNOS])$` = "paste(delta ^ \\1, \\2)", `^210Pb$` =
"paste({}^210, Pb)", `^Pb210$` = "paste({}^210, Pb)"),
units = character(0),
default_units = NA_character_,
...
)
facet_geochem_gridh(
param,
grouping = NULL,
rotate_axis_labels = 90,
scales = "free_x",
space = "fixed",
labeller = label_geochem,
renamers = c(`^d([0-9]+)([HCNOS])$` = "paste(delta ^ \\1, \\2)", `^210Pb$` =
"paste({}^210, Pb)", `^Pb210$` = "paste({}^210, Pb)"),
units = character(0),
default_units = NA_character_,
...
)Arguments
- taxon, param
A call to vars, defining the column that identifies the taxon (parameter).
- grouping
A call to vars, identifying additional grouping columns
- rotate_facet_labels, rotate_axis_labels
Facet (axis) label rotation (degrees)
- labeller
Labeller to process facet names. Use label_species to italicize species names, label_geochem to perform common formatting and units, or label_value to suppress.
- space, scales
Modify default scale freedom behaviour
- dont_italicize
Regular expressions that should not be italicized
- ...
Passed to facet_grid (abundance) or facet_wrap (geochem).
- renamers
Search and replace operations to perform in the form search = replace. Replace text can (should) contain backreferences, and will be parsed as an expression (see plotmath). Use NULL to suppress renaming.
- units
A named list of values = unit
- default_units
The default units to apply
Value
A subclass of ggplot2::facet_grid() or ggplot2::facet_wrap().
Examples
library(ggplot2)
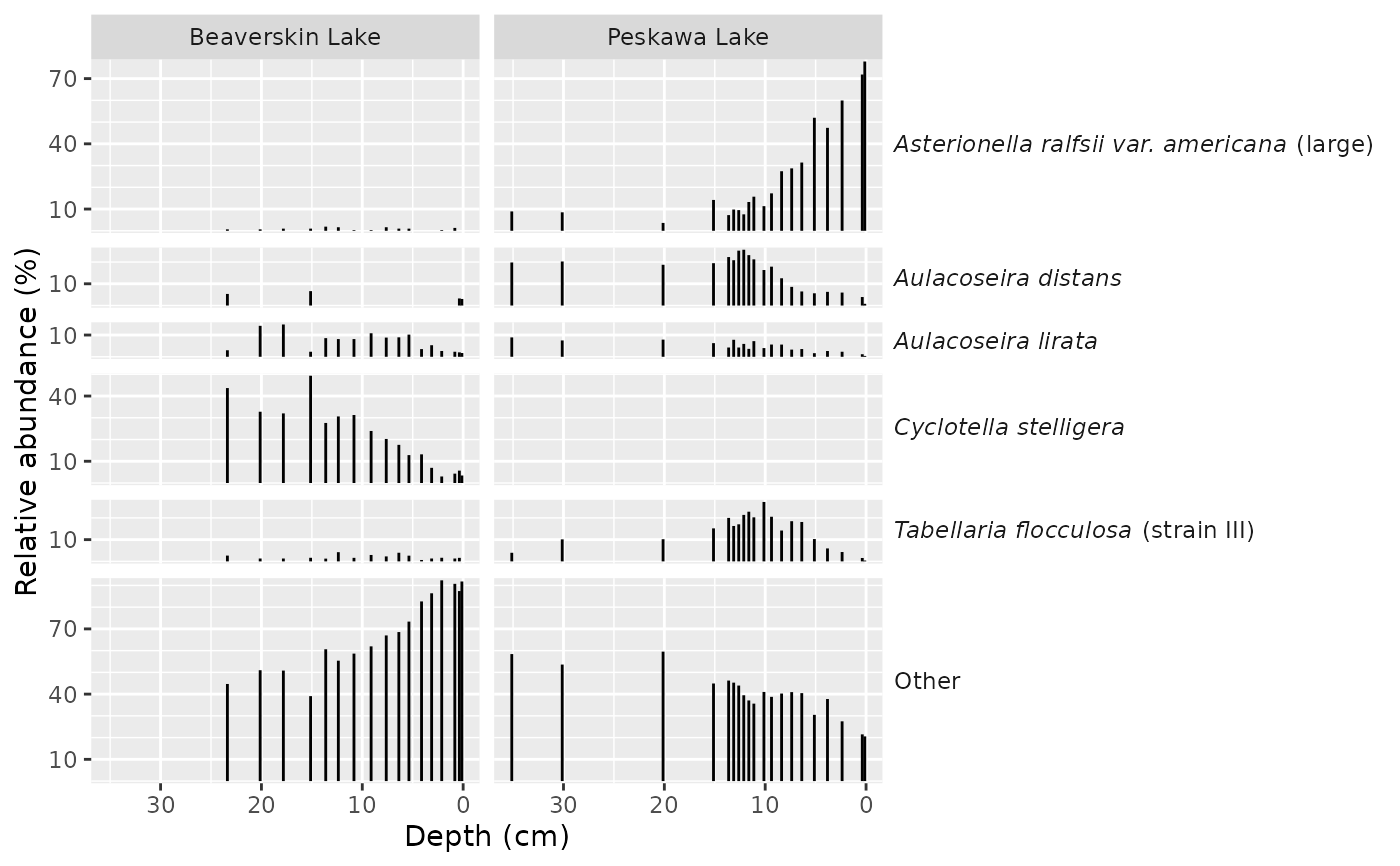
ggplot(keji_lakes_plottable, aes(x = rel_abund, y = depth)) +
geom_col_segsh() +
scale_y_reverse() +
facet_abundanceh(vars(taxon), grouping = vars(location)) +
labs(y = "Depth (cm)")
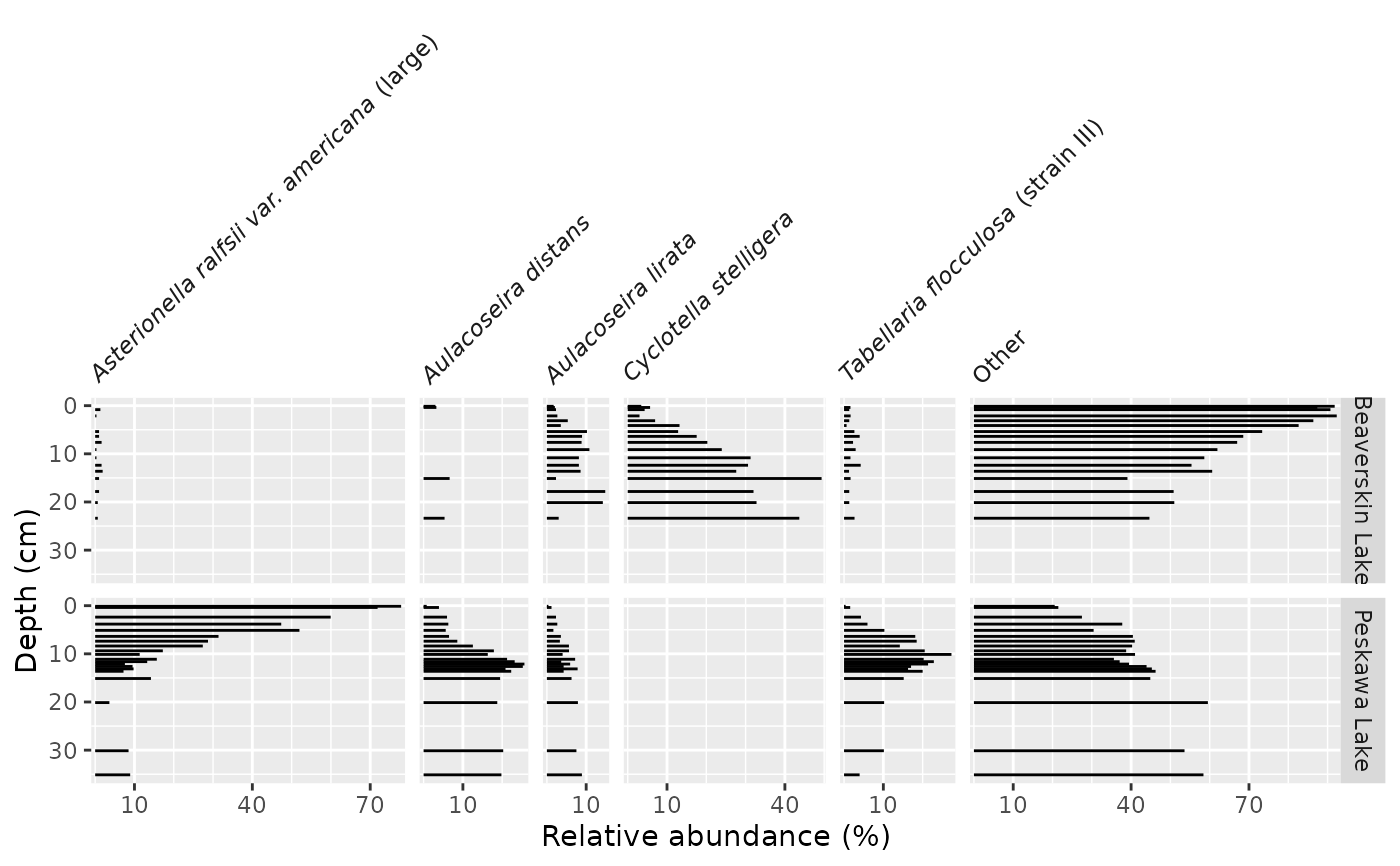 ggplot(keji_lakes_plottable, aes(y = rel_abund, x = depth)) +
geom_col_segs() +
scale_x_reverse() +
facet_abundance(vars(taxon), grouping = vars(location)) +
labs(x = "Depth (cm)")
ggplot(keji_lakes_plottable, aes(y = rel_abund, x = depth)) +
geom_col_segs() +
scale_x_reverse() +
facet_abundance(vars(taxon), grouping = vars(location)) +
labs(x = "Depth (cm)")
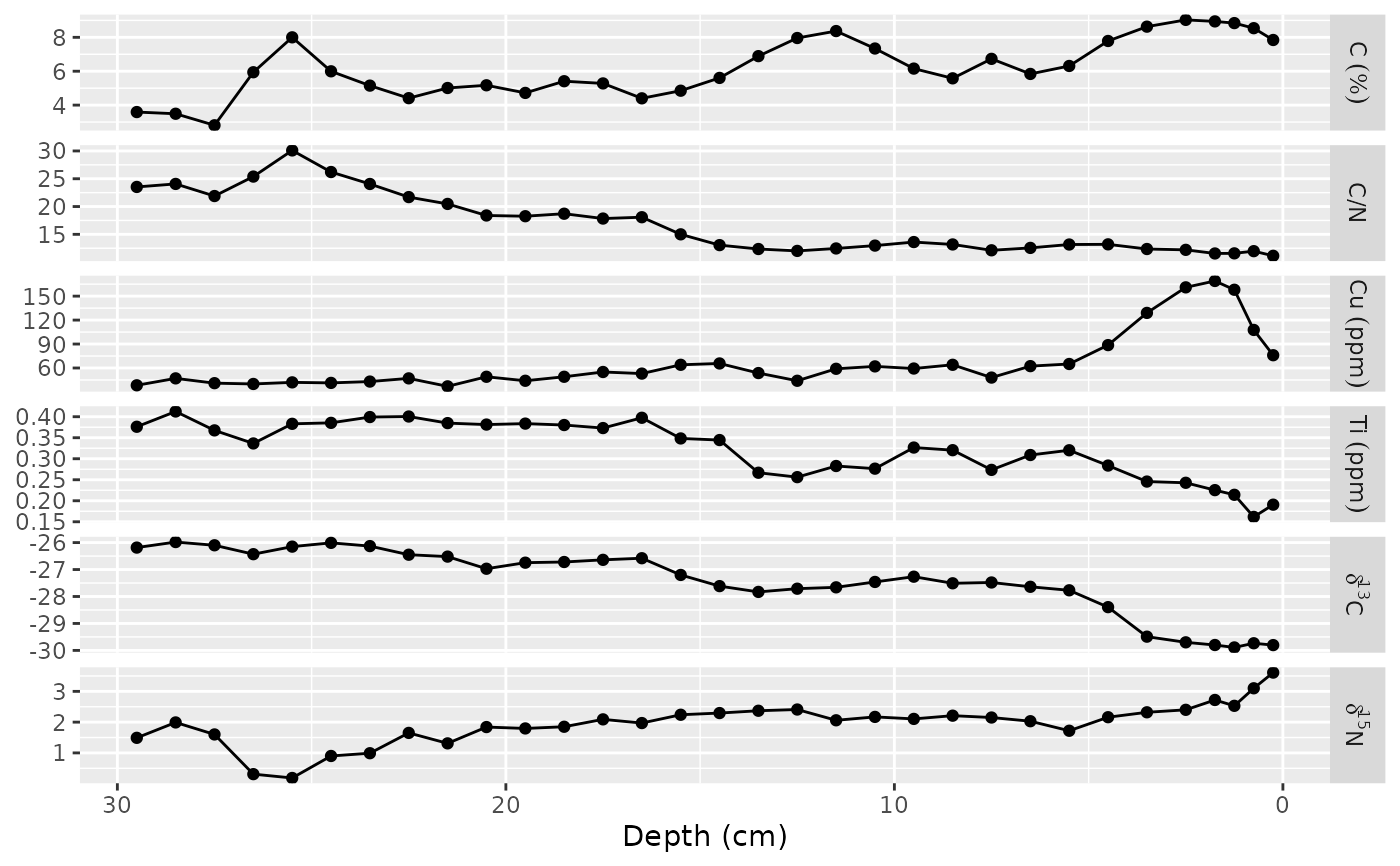
 ggplot(alta_lake_geochem, aes(x = value, y = depth)) +
geom_lineh() +
geom_point() +
scale_y_reverse() +
facet_geochem_wrap(vars(param), units = c(C = "%", Cu = "ppm", Ti = "ppm"), nrow = 1) +
labs(x = NULL, y = "Depth (cm)")
ggplot(alta_lake_geochem, aes(x = value, y = depth)) +
geom_lineh() +
geom_point() +
scale_y_reverse() +
facet_geochem_wrap(vars(param), units = c(C = "%", Cu = "ppm", Ti = "ppm"), nrow = 1) +
labs(x = NULL, y = "Depth (cm)")
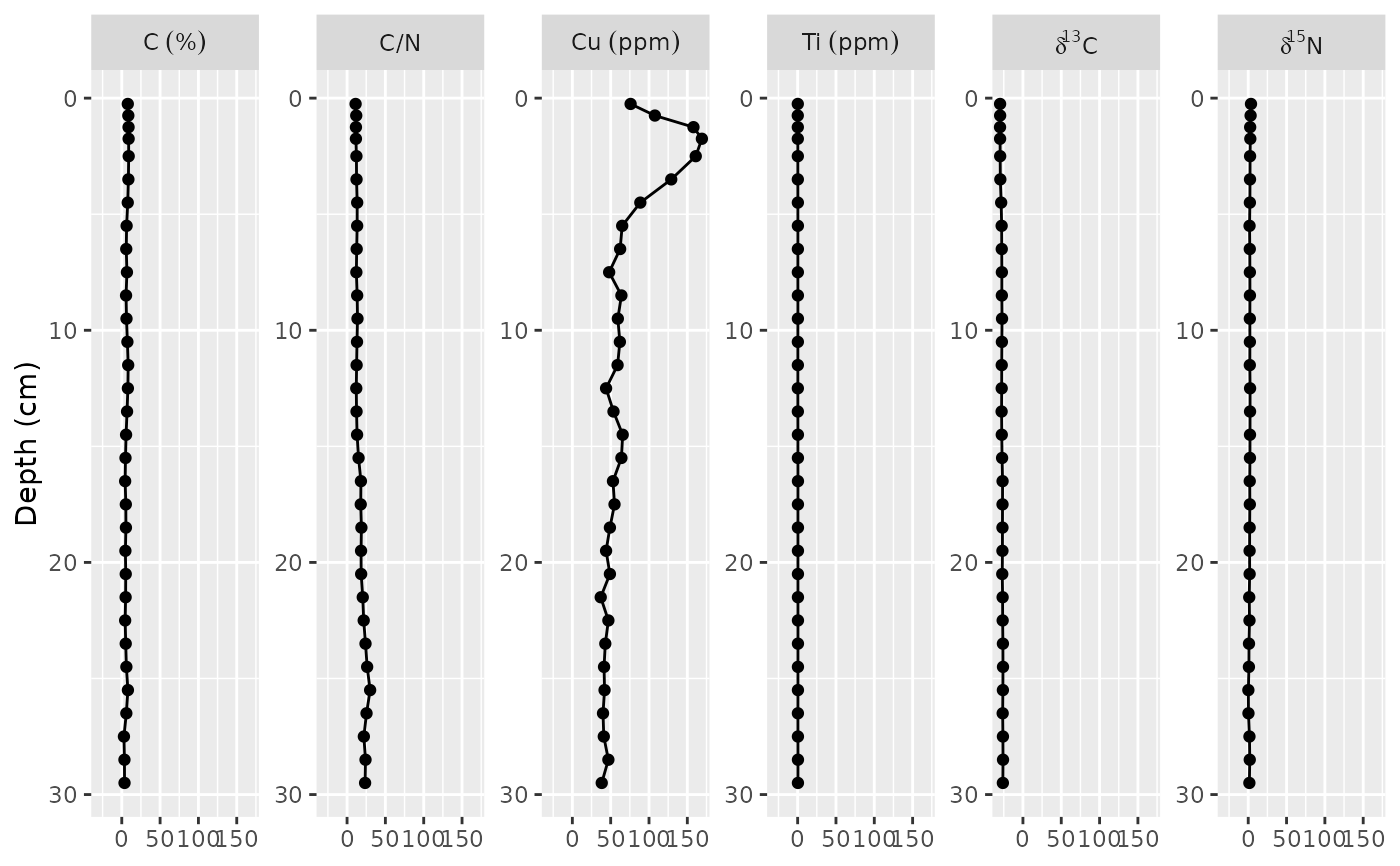 ggplot(alta_lake_geochem, aes(x = value, y = depth)) +
geom_lineh() +
geom_point() +
scale_y_reverse() +
facet_geochem_gridh(vars(param), units = c(C = "%", Cu = "ppm", Ti = "ppm")) +
labs(x = NULL, y = "Depth (cm)")
ggplot(alta_lake_geochem, aes(x = value, y = depth)) +
geom_lineh() +
geom_point() +
scale_y_reverse() +
facet_geochem_gridh(vars(param), units = c(C = "%", Cu = "ppm", Ti = "ppm")) +
labs(x = NULL, y = "Depth (cm)")
 ggplot(alta_lake_geochem, aes(y = value, x = depth)) +
geom_line() +
geom_point() +
scale_x_reverse() +
facet_geochem_grid(vars(param), units = c(C = "%", Cu = "ppm", Ti = "ppm")) +
labs(y = NULL, x = "Depth (cm)")
ggplot(alta_lake_geochem, aes(y = value, x = depth)) +
geom_line() +
geom_point() +
scale_x_reverse() +
facet_geochem_grid(vars(param), units = c(C = "%", Cu = "ppm", Ti = "ppm")) +
labs(y = NULL, x = "Depth (cm)")