nested_hclust(
.data,
data_column = "data",
qualifiers_column = "qualifiers",
distance_fun = stats::dist,
n_groups = NULL,
...,
.fun = stats::hclust,
.reserved_names = character(0)
)
nested_chclust_conslink(
.data,
data_column = "data",
qualifiers_column = "qualifiers",
distance_fun = stats::dist,
n_groups = NULL,
...
)
nested_chclust_coniss(
.data,
data_column = "data",
qualifiers_column = "qualifiers",
distance_fun = stats::dist,
n_groups = NULL,
...
)Arguments
- .data
A data frame with a list column of data frames, possibly created using nested_data.
- data_column
An expression that evalulates to the data object within each row of .data
- qualifiers_column
The column that contains the qualifiers
- distance_fun
- n_groups
The number of groups to use (can be a vector or expression using vars in .data)
- ...
- .fun
Function powering the clustering. Must return an hclust object of some kind.
- .reserved_names
Names that should not be allowed as columns in any data frame within this object
Value
.data with additional columns
References
Bennett, K. (1996) Determination of the number of zones in a biostratigraphic sequence. New Phytologist, 132, 155-170. doi:10.1111/j.1469-8137.1996.tb04521.x (Broken stick)
Grimm, E.C. (1987) CONISS: A FORTRAN 77 program for stratigraphically constrained cluster analysis by the method of incremental sum of squares. Computers & Geosciences, 13, 13-35. doi:10.1016/0098-3004(87)90022-7
Juggins, S. (2017) rioja: Analysis of Quaternary Science Data, R package version (0.9-15.1). (https://cran.r-project.org/package=rioja).
See hclust for hierarchical clustering references
Examples
library(tidyr)
library(dplyr, warn.conflicts = FALSE)
nested_coniss <- keji_lakes_plottable %>%
group_by(location) %>%
nested_data(depth, taxon, rel_abund, fill = 0) %>%
nested_chclust_coniss()
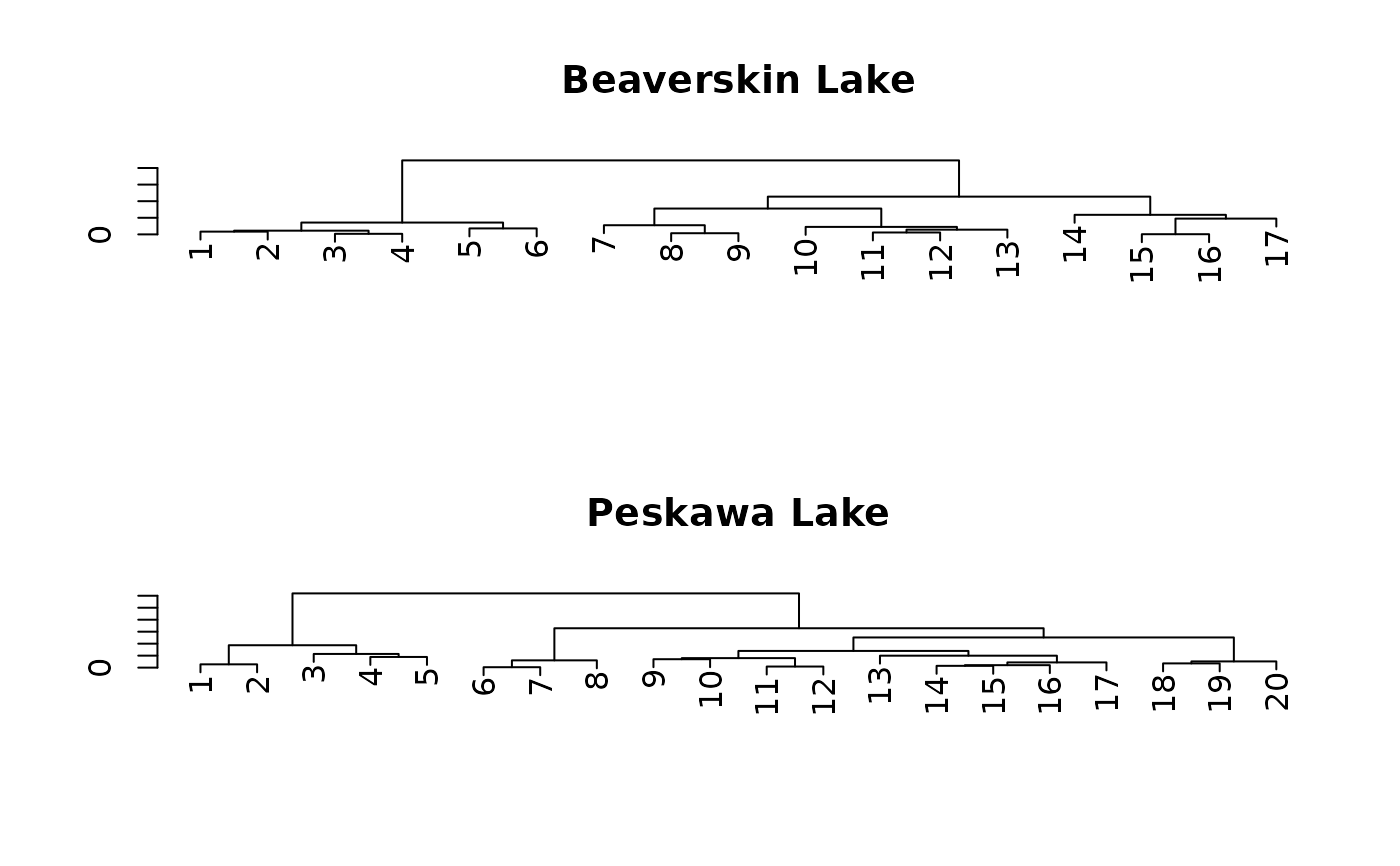
# plot the dendrograms using base graphics
plot(nested_coniss, main = location, ncol = 1)
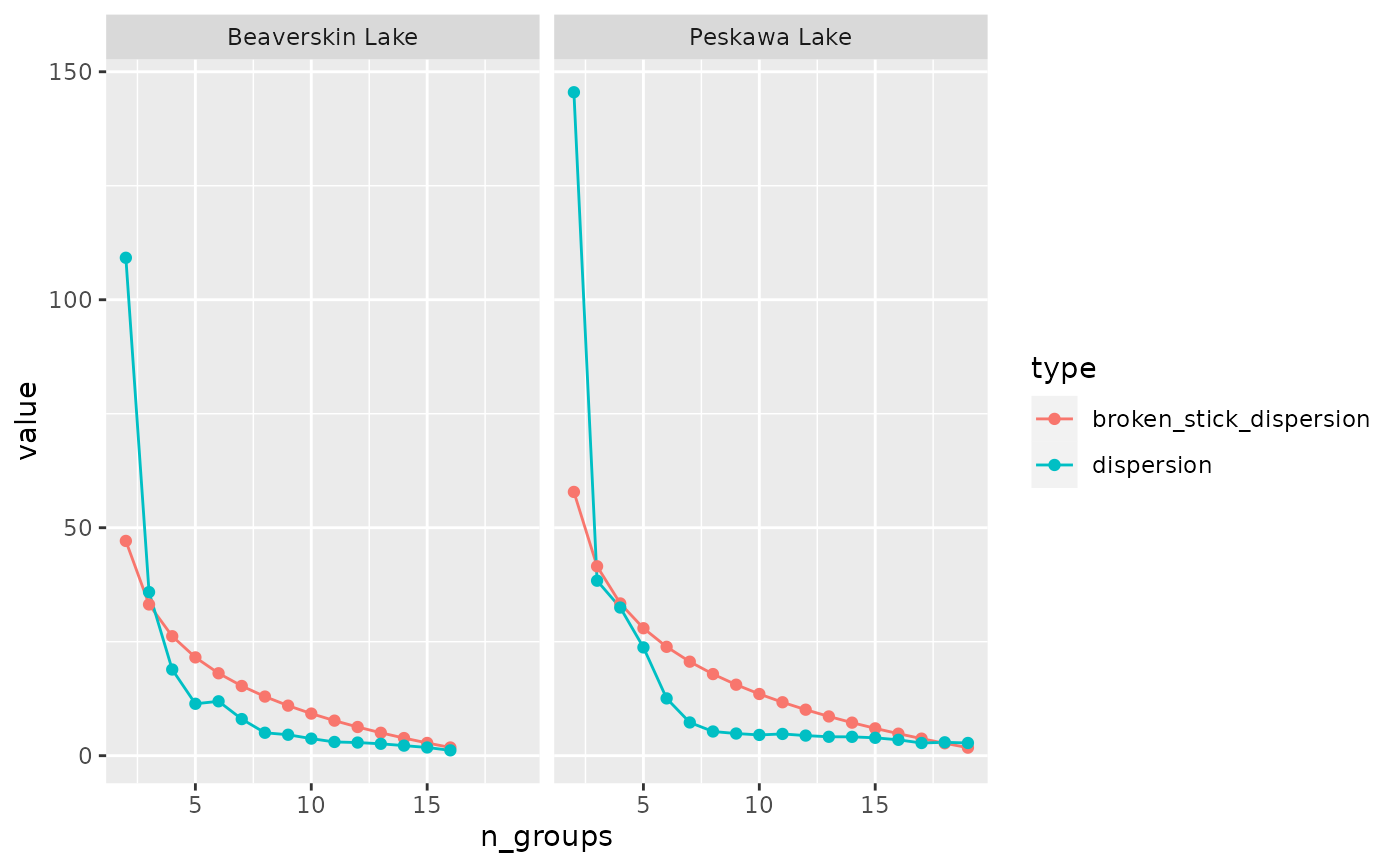
 # plot broken stick dispersion to verify number of plausible groups
library(ggplot2)
nested_coniss %>%
select(location, broken_stick) %>%
unnest(broken_stick) %>%
tidyr::gather(type, value, broken_stick_dispersion, dispersion) %>%
ggplot(aes(x = n_groups, y = value, col = type)) +
geom_line() +
geom_point() +
facet_wrap(vars(location))
# plot broken stick dispersion to verify number of plausible groups
library(ggplot2)
nested_coniss %>%
select(location, broken_stick) %>%
unnest(broken_stick) %>%
tidyr::gather(type, value, broken_stick_dispersion, dispersion) %>%
ggplot(aes(x = n_groups, y = value, col = type)) +
geom_line() +
geom_point() +
facet_wrap(vars(location))