Tutorial 4 Preparing and Loading Data
Intro
Draws from data transformation, tidy data and data import from R for Data Science (Grolemund and Wickham 2017).
4.1 Prerequisites
The prerequisites for this tutorial are tidyverse. If this package isn’t installed, you’ll have to install it using install.packages().
Load the package when you’re done! If there are errors, you may have not installed the above packages correctly!
Finally, you will need to obtain the example data. In this tutorial, we will use Lake Arnold diatom counts (Whitehead et al. 1989), obtained from the Neotoma database. You will need to download the tidy CSV version of the data, and the Excel version of the data.
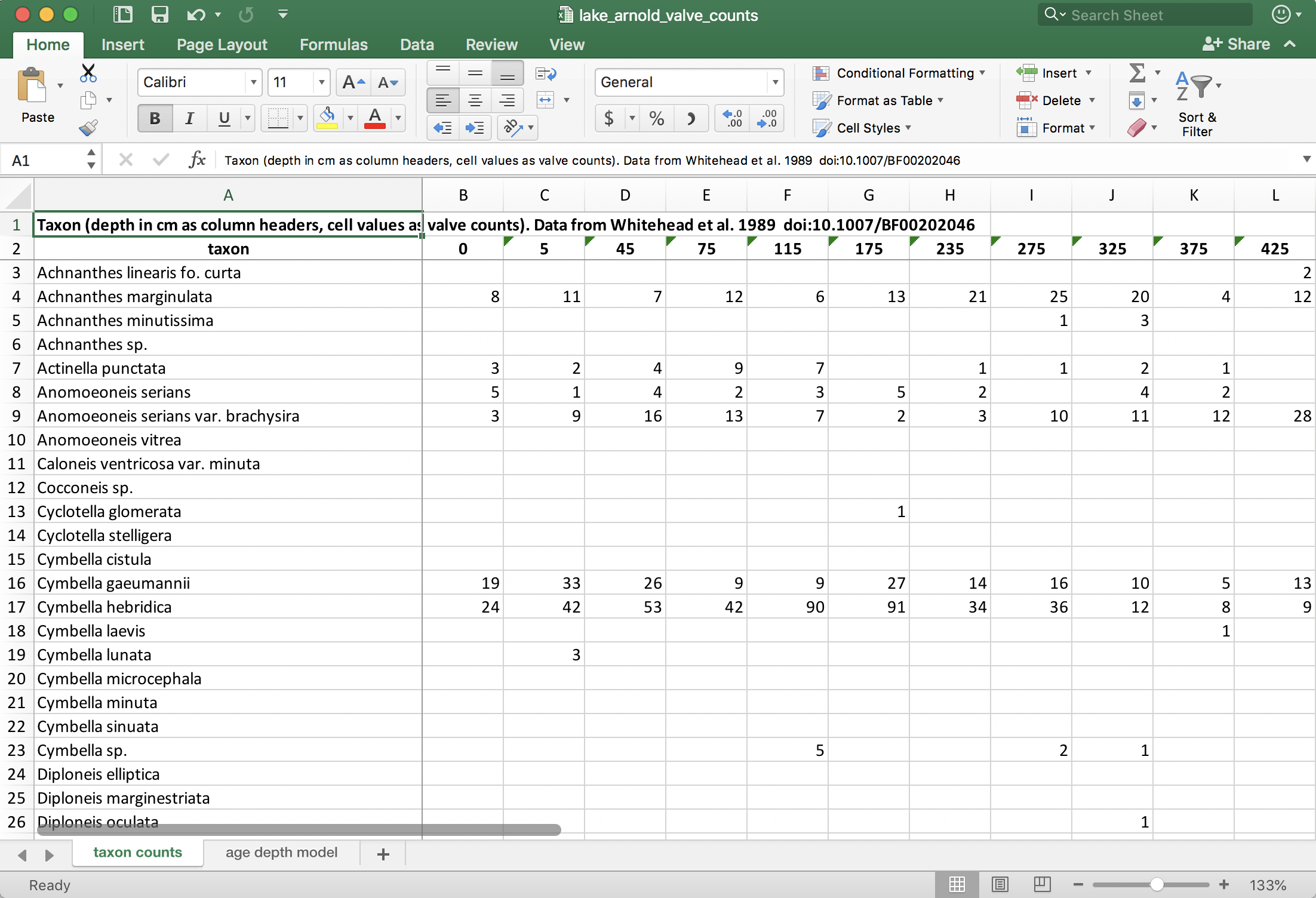
Figure 4.1: Excel screenshot of the Lake Arnold data. The format of one row per taxon with one column per sample is one way that these data are organized in the wild.
4.2 Loading data
If data are in a CSV, use read_csv():
## Parsed with column specification:
## cols(
## .default = col_integer()
## )## See spec(...) for full column specifications.The function will guess all the column types, which you can copy and paste into your read call like this:
arnold_counts_csv <- read_csv(
"data/lake_arnold_valve_counts_tidy.csv",
col_types = cols(
.default = col_integer()
)
)This will make sure that your column types are read properly in the future, and if any columns are guessed incorrectly, they can be fixed here. For more see the data import chapter of Grolemund and Wickham (2017).
We will revisit the arnold_counts_csv data frame shortly. It contains the data in the form expected by many R functions, particularly for ordination/transfer function calculations, however it is more common to see the data in a form shown in Figure 4.1. If data are in an Excel sheet, we can use the readxl package. This package is installed with the tidyverse, but you need to load it explicitly to use the read_excel() function.
4.3 Transforming data
gather():
## # A tibble: 2,128 x 3
## taxon depth_cm valve_count
## <chr> <chr> <dbl>
## 1 Achnanthes linearis fo. curta 0 NA
## 2 Achnanthes marginulata 0 8
## 3 Achnanthes minutissima 0 NA
## 4 Achnanthes sp. 0 NA
## 5 Actinella punctata 0 3
## 6 Anomoeoneis serians 0 5
## 7 Anomoeoneis serians var. brachysira 0 3
## 8 Anomoeoneis vitrea 0 NA
## 9 Caloneis ventricosa var. minuta 0 NA
## 10 Cocconeis sp. 0 NA
## # ... with 2,118 more rowsChange type of depth column:
arnold_counts_excel %>%
gather(key = depth_cm, value = valve_count, -taxon) %>%
mutate(depth_cm = as.numeric(depth_cm))## # A tibble: 2,128 x 3
## taxon depth_cm valve_count
## <chr> <dbl> <dbl>
## 1 Achnanthes linearis fo. curta 0 NA
## 2 Achnanthes marginulata 0 8
## 3 Achnanthes minutissima 0 NA
## 4 Achnanthes sp. 0 NA
## 5 Actinella punctata 0 3
## 6 Anomoeoneis serians 0 5
## 7 Anomoeoneis serians var. brachysira 0 3
## 8 Anomoeoneis vitrea 0 NA
## 9 Caloneis ventricosa var. minuta 0 NA
## 10 Cocconeis sp. 0 NA
## # ... with 2,118 more rowsChange NA valve counts to 0:
arnold_counts_excel %>%
gather(key = depth_cm, value = valve_count, -taxon) %>%
mutate(depth_cm = as.numeric(depth_cm)) %>%
mutate(valve_count = if_else(is.na(valve_count), 0, valve_count))## # A tibble: 2,128 x 3
## taxon depth_cm valve_count
## <chr> <dbl> <dbl>
## 1 Achnanthes linearis fo. curta 0 0
## 2 Achnanthes marginulata 0 8
## 3 Achnanthes minutissima 0 0
## 4 Achnanthes sp. 0 0
## 5 Actinella punctata 0 3
## 6 Anomoeoneis serians 0 5
## 7 Anomoeoneis serians var. brachysira 0 3
## 8 Anomoeoneis vitrea 0 0
## 9 Caloneis ventricosa var. minuta 0 0
## 10 Cocconeis sp. 0 0
## # ... with 2,118 more rowsThis form of data is particularly useful because it contains one row per measurement, and looses no data. When finished constructing the pipe, assign to a variable:
arnold_counts <- arnold_counts_excel %>%
gather(key = depth_cm, value = valve_count, -taxon) %>%
mutate(depth_cm = as.numeric(depth_cm)) %>%
mutate(valve_count = if_else(is.na(valve_count), 0, valve_count))Things we can do with the data in this form:
Use a grouped mutate to calculate relative abundance:
arnold_rel_abund <- arnold_counts %>%
group_by(depth_cm) %>%
mutate(relative_abundance_percent = valve_count / sum(valve_count) * 100) %>%
ungroup()
arnold_rel_abund## # A tibble: 2,128 x 4
## taxon depth_cm valve_count relative_abundance_per…
## <chr> <dbl> <dbl> <dbl>
## 1 Achnanthes linearis fo. c… 0 0 0
## 2 Achnanthes marginulata 0 8 2.07
## 3 Achnanthes minutissima 0 0 0
## 4 Achnanthes sp. 0 0 0
## 5 Actinella punctata 0 3 0.775
## 6 Anomoeoneis serians 0 5 1.29
## 7 Anomoeoneis serians var. … 0 3 0.775
## 8 Anomoeoneis vitrea 0 0 0
## 9 Caloneis ventricosa var. … 0 0 0
## 10 Cocconeis sp. 0 0 0
## # ... with 2,118 more rowsSummarise by taxon, find most common taxa:
arnold_rel_abund %>%
group_by(taxon) %>%
summarise(
min_rel_abund = min(relative_abundance_percent),
max_rel_abund = max(relative_abundance_percent),
mean_rel_abund = mean(relative_abundance_percent)
) %>%
arrange(desc(max_rel_abund))## # A tibble: 133 x 4
## taxon min_rel_abund max_rel_abund mean_rel_abund
## <chr> <dbl> <dbl> <dbl>
## 1 Fragilaria construens 0 30.8 1.92
## 2 Fragilaria virescens var. b… 0 25.6 3.08
## 3 Cymbella hebridica 0 23.7 7.21
## 4 Melosira distans 0 23.5 7.18
## 5 Pinnularia biceps 0 21.1 7.13
## 6 Pinnularia microstauron 0 20.6 2.68
## 7 Fragilaria brevistriata 0 15.2 0.953
## 8 Fragilaria pinnata 0 15 0.954
## 9 Navicula subtilissima 0 14.9 6.21
## 10 Anomoeoneis serians var. br… 0 13.0 4.78
## # ... with 123 more rowsSummarise by depth, find depths with greatest richness:
arnold_rel_abund %>%
group_by(depth_cm) %>%
summarise(
n_valves = sum(valve_count),
total_rel_abund = sum(relative_abundance_percent),
n_taxa = sum(valve_count > 0)
)## # A tibble: 16 x 4
## depth_cm n_valves total_rel_abund n_taxa
## <dbl> <dbl> <dbl> <int>
## 1 0 387 100 44
## 2 5 489 100 50
## 3 45 398 100 51
## 4 75 368 100 40
## 5 115 484 100 44
## 6 175 384 100 51
## 7 235 346 100 47
## 8 275 381 100 47
## 9 325 339 100 50
## 10 375 394 100 40
## 11 425 245 100 36
## 12 445 382 100 44
## 13 475 386 100 44
## 14 495 384 100 47
## 15 525 375 100 42
## 16 555 400 100 29Create form of data with one row per depth value (this is almost the same as the arnold_counts_csv that we loaded earlier):
arnold_rel_abund %>%
select(taxon, depth_cm, relative_abundance_percent) %>%
spread(key = taxon, value = relative_abundance_percent)## # A tibble: 16 x 134
## depth_cm `Achnanthes linearis … `Achnanthes margin… `Achnanthes minuti…
## <dbl> <dbl> <dbl> <dbl>
## 1 0 0 2.07 0
## 2 5 0 2.25 0
## 3 45 0 1.76 0
## 4 75 0 3.26 0
## 5 115 0 1.24 0
## 6 175 0 3.39 0
## 7 235 0 6.07 0
## 8 275 0 6.56 0.262
## 9 325 0 5.90 0.885
## 10 375 0 1.02 0
## 11 425 0.816 4.90 0
## 12 445 1.57 3.93 0
## 13 475 0 6.22 0
## 14 495 0 3.12 0
## 15 525 0 1.33 5.6
## 16 555 0 0 0
## # ... with 130 more variables: `Achnanthes sp.` <dbl>, `Actinella
## # punctata` <dbl>, `Anomoeoneis serians` <dbl>, `Anomoeoneis serians
## # var. brachysira` <dbl>, `Anomoeoneis vitrea` <dbl>, `Caloneis
## # ventricosa var. minuta` <dbl>, `Cocconeis sp.` <dbl>, `Cyclotella
## # glomerata` <dbl>, `Cyclotella stelligera` <dbl>, `Cymbella
## # cistula` <dbl>, `Cymbella gaeumannii` <dbl>, `Cymbella
## # hebridica` <dbl>, `Cymbella laevis` <dbl>, `Cymbella lunata` <dbl>,
## # `Cymbella microcephala` <dbl>, `Cymbella minuta` <dbl>, `Cymbella
## # sinuata` <dbl>, `Cymbella sp.` <dbl>, `Diploneis elliptica` <dbl>,
## # `Diploneis marginestriata` <dbl>, `Diploneis oculata` <dbl>, `Eunotia
## # arcus` <dbl>, `Eunotia assymetrica` <dbl>, `Eunotia bactriana` <dbl>,
## # `Eunotia bidentula` <dbl>, `Eunotia bigibba` <dbl>, `Eunotia
## # curvata` <dbl>, `Eunotia diodon` <dbl>, `Eunotia elegans` <dbl>,
## # `Eunotia exigua` <dbl>, `Eunotia fallax` <dbl>, `Eunotia
## # flexuosa` <dbl>, `Eunotia gibbosa` <dbl>, `Eunotia hexaglyphis` <dbl>,
## # `Eunotia incisa` <dbl>, `Eunotia major` <dbl>, `Eunotia
## # microcephala` <dbl>, `Eunotia monodon` <dbl>, `Eunotia
## # naegelii` <dbl>, `Eunotia pectinalis` <dbl>, `Eunotia pectinalis var.
## # minor` <dbl>, `Eunotia perpusilla` <dbl>, `Eunotia praerupta` <dbl>,
## # `Eunotia serra` <dbl>, `Eunotia soleirolii` <dbl>, `Eunotia
## # spp.` <dbl>, `Eunotia sudetica` <dbl>, `Eunotia suecica` <dbl>,
## # `Eunotia tenella` <dbl>, `Eunotia triodon` <dbl>, `Eunotia
## # vanheurckii` <dbl>, `Eunotia vanheurckii var. intermedia` <dbl>,
## # `Fragilaria brevistriata` <dbl>, `Fragilaria constricta` <dbl>,
## # `Fragilaria constricta fo. stricta` <dbl>, `Fragilaria
## # construens` <dbl>, `Fragilaria construens var. binodis` <dbl>,
## # `Fragilaria construens var. pumila` <dbl>, `Fragilaria construens var.
## # venter` <dbl>, `Fragilaria pinnata` <dbl>, `Fragilaria sp.` <dbl>,
## # `Fragilaria virescens` <dbl>, `Fragilaria virescens var.
## # birostrata` <dbl>, `Frustulia rhomboides` <dbl>, `Frustulia rhomboides
## # var. capitata` <dbl>, `Frustulia rhomboides var. crassinervia` <dbl>,
## # `Frustulia rhomboides var. saxonica` <dbl>, `Gomphonema
## # angustatum` <dbl>, `Gomphonema gracile` <dbl>, `Gomphonema
## # parvulum` <dbl>, `Gomphonema sp.` <dbl>, `Melosira distans` <dbl>,
## # `Melosira distans var. africana (small) SER ADIR` <dbl>, `Melosira
## # distans var. lirata` <dbl>, `Melosira distans var. lirata fo.
## # seriata` <dbl>, `Melosira italica var. subarctica` <dbl>, `Melosira
## # sp.` <dbl>, `Navicula angusta` <dbl>, `Navicula gysingensis` <dbl>,
## # `Navicula mediocris` <dbl>, `Navicula minima` <dbl>, `Navicula
## # monmouthiana-stodderi` <dbl>, `Navicula mutica` <dbl>, `Navicula
## # notha` <dbl>, `Navicula pseudoscutiformis` <dbl>, `Navicula
## # radiosa` <dbl>, `Navicula sp.` <dbl>, `Navicula subtilissima` <dbl>,
## # `Navicula tenuicephala` <dbl>, `Navicula vanheurckii` <dbl>, `Neidium
## # affine` <dbl>, `Neidium affine var. longiceps` <dbl>, `Neidium
## # bisulcatum` <dbl>, `Neidium herrmannii` <dbl>, `Neidium iridis` <dbl>,
## # `Neidium iridis var. amphigomphus` <dbl>, `Neidium sp.` <dbl>,
## # `Nitzschia acuta` <dbl>, `Nitzschia dissipata` <dbl>, `Nitzschia
## # linearis` <dbl>, …Use a left-join to add age-depth info:
arnold_age_depth <- read_excel(
"data/lake_arnold_valve_counts.xlsx",
sheet = "age depth model",
skip = 1
)
arnold_rel_abund %>%
left_join(arnold_age_depth, by = "depth_cm") %>%
select(taxon, depth_cm, age_bp, everything())## # A tibble: 2,128 x 5
## taxon depth_cm age_bp valve_count relative_abundance_p…
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Achnanthes linearis … 0 -39 0 0
## 2 Achnanthes marginula… 0 -39 8 2.07
## 3 Achnanthes minutissi… 0 -39 0 0
## 4 Achnanthes sp. 0 -39 0 0
## 5 Actinella punctata 0 -39 3 0.775
## 6 Anomoeoneis serians 0 -39 5 1.29
## 7 Anomoeoneis serians … 0 -39 3 0.775
## 8 Anomoeoneis vitrea 0 -39 0 0
## 9 Caloneis ventricosa … 0 -39 0 0
## 10 Cocconeis sp. 0 -39 0 0
## # ... with 2,118 more rowsFilter to only include some taxa:
Find common taxa:
common_taxa <- arnold_rel_abund %>%
group_by(taxon) %>%
summarise(
max_rel_abund = max(relative_abundance_percent)
) %>%
filter(max_rel_abund >= 20) %>%
pull(taxon)
common_taxa## [1] "Cymbella hebridica"
## [2] "Fragilaria construens"
## [3] "Fragilaria virescens var. birostrata"
## [4] "Melosira distans"
## [5] "Pinnularia biceps"
## [6] "Pinnularia microstauron"Filter the counts:
## # A tibble: 96 x 4
## taxon depth_cm valve_count relative_abundance_pe…
## <chr> <dbl> <dbl> <dbl>
## 1 Cymbella hebridica 0 24 6.20
## 2 Fragilaria construens 0 0 0
## 3 Fragilaria virescens var. … 0 99 25.6
## 4 Melosira distans 0 0 0
## 5 Pinnularia biceps 0 0 0
## 6 Pinnularia microstauron 0 0 0
## 7 Cymbella hebridica 5 42 8.59
## 8 Fragilaria construens 5 0 0
## 9 Fragilaria virescens var. … 5 90 18.4
## 10 Melosira distans 5 1 0.204
## # ... with 86 more rows4.4 Exporing data
- use write_csv, writexl::write_xlsx, to write output
4.5 Summary
Tutorial summary
Draws from data transformation and data import from R for Data Science.
References
Grolemund, Garrett, and Hadley Wickham. 2017. R for Data Science. New York: O’Reily. http://r4ds.had.co.nz/.
Whitehead, Donald R., Donald F. Charles, Stephen T. Jackson, John P. Smol, and Daniel R. Engstrom. 1989. “The Developmental History of Adirondack (N.Y.) Lakes.” Journal of Paleolimnology 2 (3): 185–206. https://doi.org/10.1007/BF00202046.